The goal of our laboratory is to define the molecular pathways necessary to maintain homeostasis in both the developing and aging nervous system. To do this we utilize both phenotype-driven forward genetics and gene-driven reverse genetics to identify mutations that are associated with abnormal CNS development or neurodegeneration. Our approach has uncovered disruptions in several novel pathways that were not previously associated with loss of neuronal function but have been shown to cause human disease subsequent to our studies. We also use forward genetics to identify single-locus suppressor/enhancer genes of these mutations to enhance our understanding of the molecular pathways underlying these phenotypes and our progress toward therapeutic innovation in these diseases. We are particularly interested in the role of alterations in translation elongation, translational fidelity, proteostasis, and RNA metabolism in neuronal function.
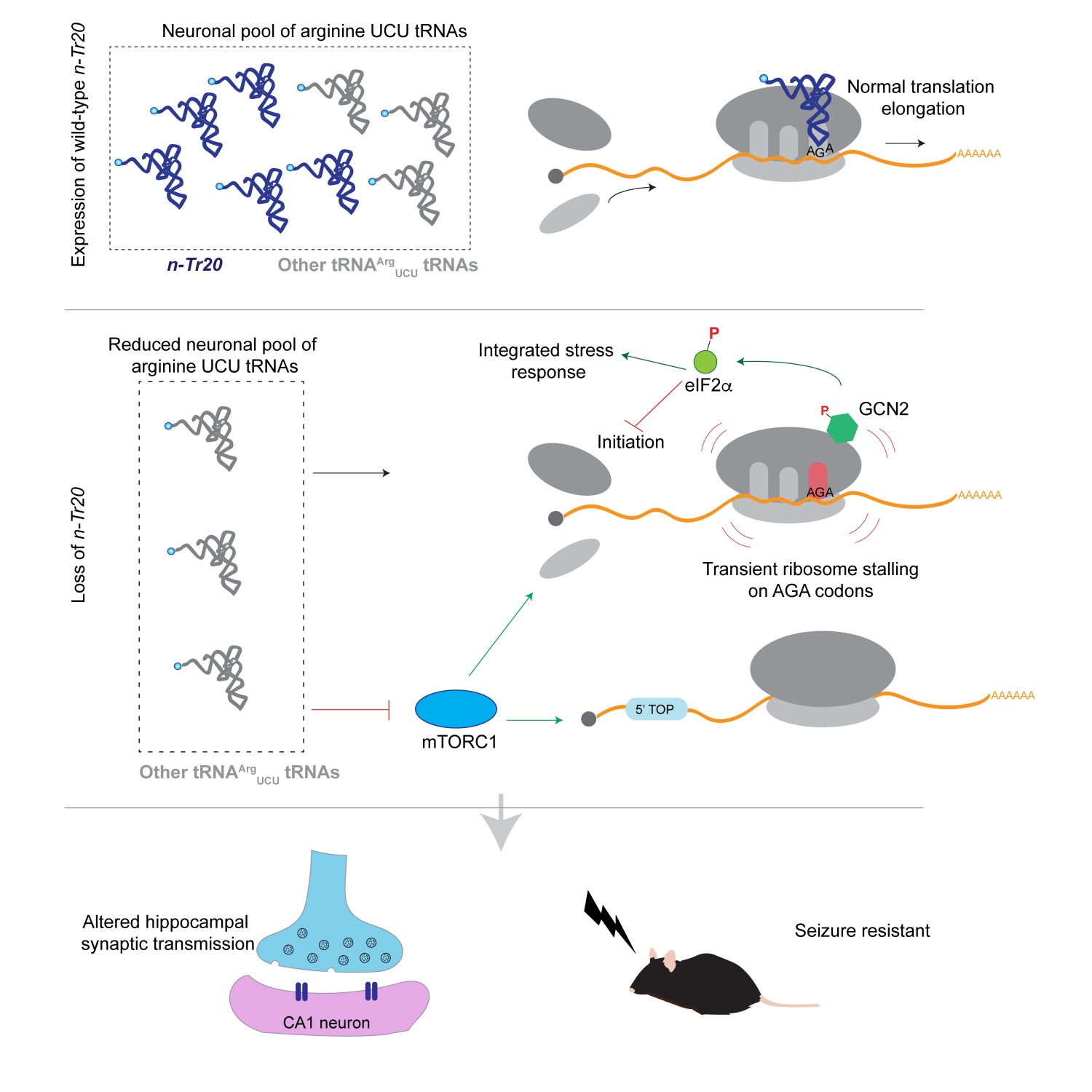
The mammalian genome has hundreds of nuclear-encoded tRNAs, but the contribution of individual tRNA genes to cellular and organismal function remains unknown. Here, we demonstrate that mutations in a neuronally enriched arginine tRNA, n-Tr20, increased seizure threshold and altered synaptic transmission. n-Tr20 expression also modulated seizures caused by an epilepsy-linked mutation in Gabrg2, a gene encoding a GABAA receptor subunit. Loss of n-Tr20 altered translation initiation by activating the integrated stress response and suppressing mTOR signaling, the latter of which may contribute to altered neurotransmission in mutant mice. Deletion of a highly expressed isoleucine tRNA similarly altered these signaling pathways in the brain, suggesting that regulation of translation initiation is a conserved response to tRNA loss. Our data indicate that loss of a single member of a tRNA family results in multiple cellular phenotypes, highlighting the disease-causing potential of tRNA mutations.


