The goal of our laboratory is to define the molecular pathways necessary to maintain homeostasis in both the developing and aging nervous system. To do this we utilize both phenotype-driven forward genetics and gene-driven reverse genetics to identify mutations that are associated with abnormal CNS development or neurodegeneration. Our approach has uncovered disruptions in several novel pathways that were not previously associated with loss of neuronal function but have been shown to cause human disease subsequent to our studies. We also use forward genetics to identify single-locus suppressor/enhancer genes of these mutations to enhance our understanding of the molecular pathways underlying these phenotypes and our progress toward therapeutic innovation in these diseases. We are particularly interested in the role of alterations in translation elongation, translational fidelity, proteostasis, and RNA metabolism in neuronal function.
Mistranslation and neurodegeneration
Oligomerization and the formation of aggregates of misfolded proteins are common to many genetic and sporadic forms of neurodegenerative diseases. Although some of these misfolded proteins are caused by mutations directly within disease-related proteins, such as in the polyglutamine expansion diseases and some forms of familial Alzheimer’s disease, the mechanisms underlying protein misfolding in many sporadic forms of neurodegenerative diseases remain unknown. We used a forward genetic approach to identify novel genes that, when their function is disrupted, cause the accumulation of misfolded proteins in neurons prior to their death.
Purkinje cell loss in mice homozygous for the spontaneous sticky (sti) mutation is associated with accumulation of ubiquitinated proteins in the cytoplasm, endoplasmic reticulum, and nucleolus. We have determined that the sti molecular defect is a point mutation in the editing domain of alanyl tRNA synthetase (AlaRS). The aminoacyl tRNA synthetases establish the genetic code that links each amino acid to its cognate tRNA which bears the anticodon triplet of the code. The high accuracy of protein synthesis is largely attributable to the precision of these aminoacylation reactions, and much of this accuracy resides in the editing domains of these synthetases that clear misactivated amino acids or mischarged tRNAs. We demonstrated that the sti mutation causes an increase in mischarged serine-tRNAAla. This likely leads to random misincorporation of serine at alanine codons, ultimately causing production of heterogeneous, unfolded proteins. The loss of translational fidelity in sti mutant mice is an exciting new mechanism underlying neurodegeneration.
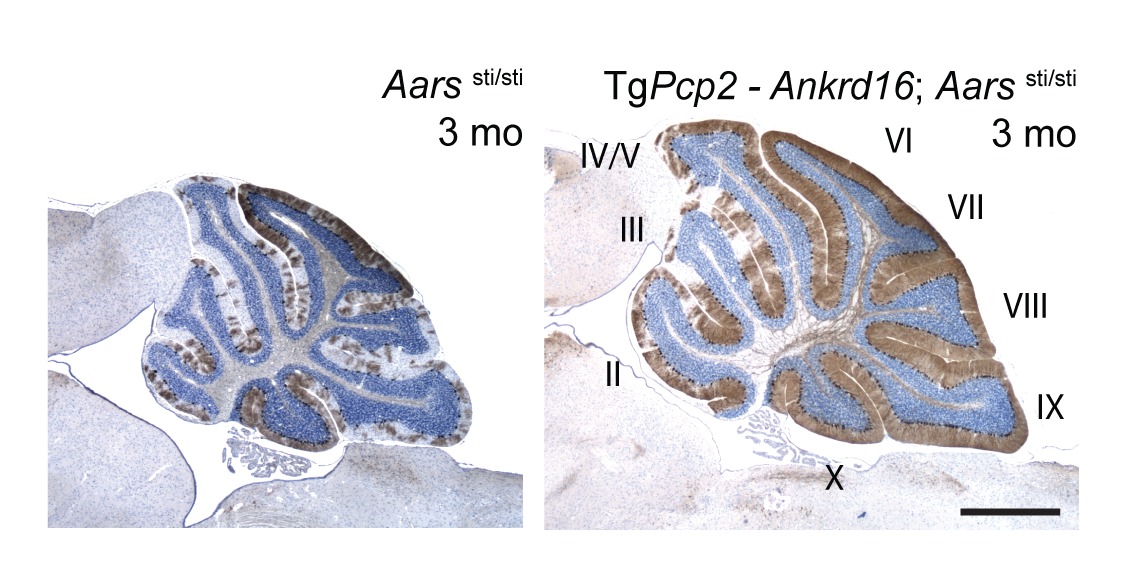
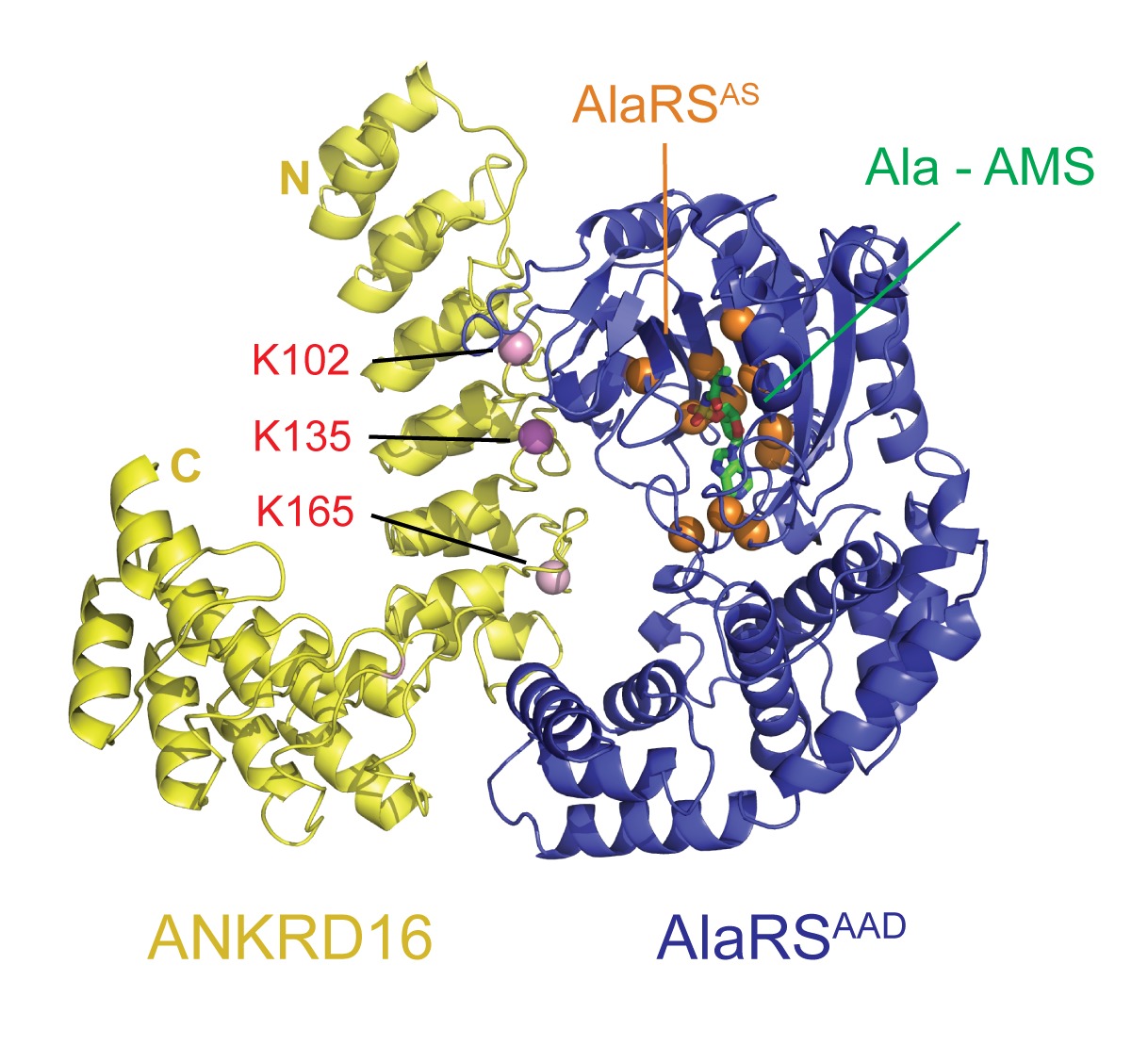
Like the mutation of Gtpbp2, Purkinje cell degeneration associated with the sti mutation varies with genetic background. Using this variation and positional cloning, we have identified the ankyrin repeat protein, ANKRD16, as a genetic modifier of sti that suppresses protein inclusion formation and neurodegeneration in mutant Purkinje cells in a gene dosage–dependent manner. Our experiments demonstrate that ANKRD16 directly interacts with AlaRS and misactivated serine is transferred to lysines on ANKRD16, thereby preventing misacylation of tRNAAla and the production of unfolded proteins. ANKRD16 is ubiquitously expressed but has lower levels in Purkinje cells suggesting that these neurons have a lower threshold for AlaRS-associated editing defects. Indeed, when Ankrd16 is deleted in the hippocampus in sti mutant mice these neurons develop protein aggregates and degenerate. We are now using various Cre lines to delete Ankrd16 in sti mutant mice to examine the effects of mistranslation on protein aggregate formation in different neuronal populations.
Work on these projects, as well as investigation of other genes identified in our screen, should continue to yield exciting insights into the mechanisms underlying neurodegeneration
Ribosome stalling and neurological disease
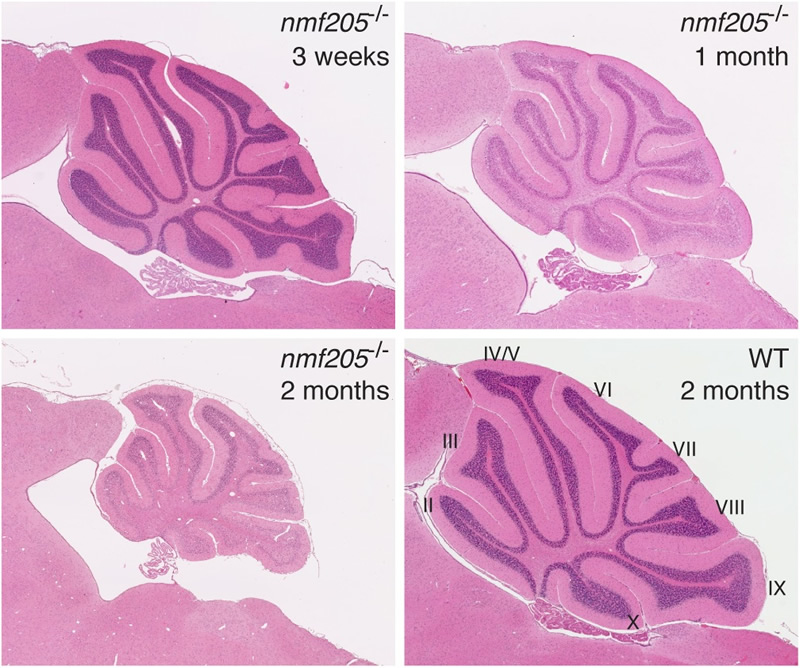
The speed of translation across a transcript can vary and contribute to correct folding of the nascent peptide. Our work has demonstrated that prolonged pauses of ribosomes during translation elongation can be detrimental for neuronal fitness. Cerebellar, hippocampal, cortical, and retinal neurons degenerate in mice homozygous for the ENU-induced mutation nmf205. We determined that these mice have a null mutation in the gene encoding GTPBP2, a novel translational GTPase.
The neurodegeneration associated with loss of Gtpbp2 function varies tremendously depending on genetic background, suggesting the presence of genes that modify this phenotype. By positional cloning, we identified a mutation in an arginine tRNA gene, n-Tr20, that results in neuron death when combined with the Gtpbp2 mutation. Surprisingly, this tRNA gene is expressed only in the nervous system. Although five tRNA genes with the same anticodon are present in the mouse genome, ribosome profiling demonstrated that loss of this isodecoder tRNA resulted in increased ribosome pausing during translation elongation when the codon complementary to the anticodon of the mutated tRNA was present in the ribosomal A site. Ribosome occupancy at this codon dramatically increased when Gtpbp2 and the tRNA gene were mutated, suggesting that GTPBP2 is a newly identified ribosome rescue factor. We are actively investigating the function of GTPBP2 and other putative ribosome quality control factors in the nervous system and searching for additional modifiers of these genes.
tRNAs, synaptic physiology, and seizure threshold
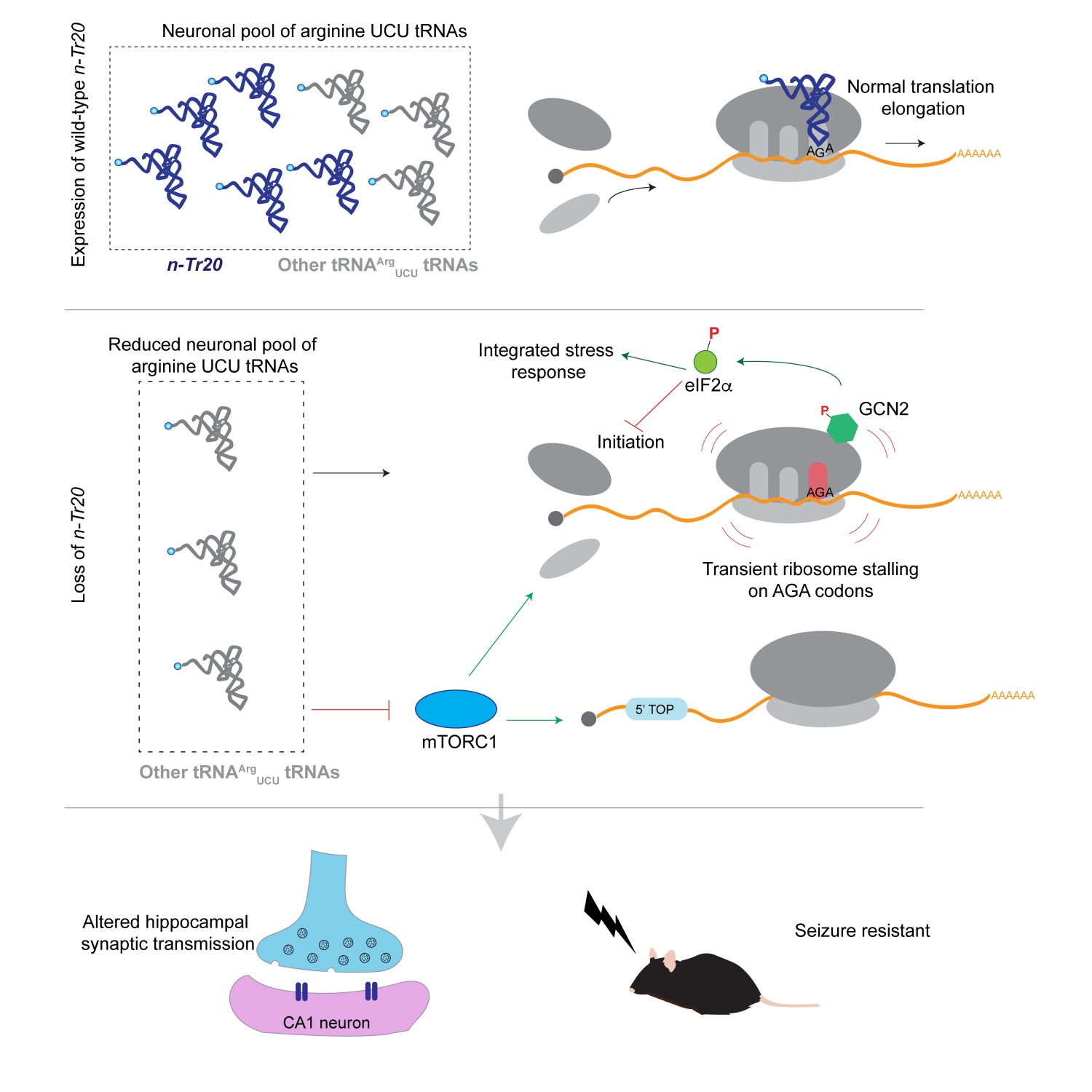
The mammalian genome has hundreds of nuclear-encoded tRNAs suggesting functional redundancy. Thus, the disease-causing potential of these genes have been largely overlooked. Interestingly the arginine UCU tRNA gene, n-Tr20, resides in a quantitative trait locus (QTL) controlling seizure threshold in inbred strains of mice. C57BL/6J (B6J) mice have a high seizure threshold relative to most inbred strains. Given that we previously showed that B6J mice have a mutation in n-Tr20 that impairs its processing, dramatically lowering the level of mature arginine UCU tRNAs in the brain, we asked whether levels of n-Tr20 regulate seizure threshold. Indeed, restoration of wild type levels of n-Tr20 in B6J mice lowered seizure threshold, and complete deletion of n-Tr20 in C57BL/6N (B6N) mice increased seizure threshold. n-Tr20 levels altered synaptic transmission and the excitatory/inhibitory balance in the hippocampus, and also modulated spike-wave discharges in a mouse model of human epilepsy.
Loss of n-Tr20 altered translation initiation by activating the integrated stress response and suppressing mTOR signaling, and rapamycin treatment of wild type (B6N) mice resulted in similar synaptic changes as deletion of n-Tr20. Similar alterations of these signaling pathways were observed upon deletion of a highly expressed isoleucine tRNA suggesting that these changes are a conserved response to tRNA loss. Currently we are extending our analysis of potential disease-causing mutations in tRNAs, the cell specific expression of tRNAs, and the mechanisms underlying synaptic changes upon loss of n-Tr20.